This is disgusting. Notice the kernel version 😎 #linux.
Nicholas M Glykos' blog
Thursday, April 24, 2025
Sunday, January 28, 2024
Can you make a useful student terminal using a Raspberry Pi 5 ?
Executive summary
Yes, you can. And not only it is doable, but it is actually good fun! The Pi’s are fully useable as student terminals, allowing students to do much more than just ssh-ing to the servers : they have a fast and responsive local environment with chromium, libreoffice, molecular graphics, any programming language they want, R & R-studio, etc. Having said that, the students using them must not be in fear of doing some system administration now and then (but with a carefully prepared image for the microSD card, even this will be very rare). All-in-all, this is a loud and clear go from my side (and at the present time).
 |
| The Pi at full throttle. Overview of four workspaces shown with molecular graphics, libreoffice writer, a tmux session, and a browser. |
The long story
Introduction
So, I decided to buy a Raspberry Pi 5. Not sure why. Probably to have a new toy to play with during the Christmas vacation. This turned-out to be a good choice: the Pi is very good fun, and I still enjoy fiddling with its settings and making it look like a proper linux machine. There were some minor issues, like a difficulty in finding suitable adaptor for micro HDMI, or -the several- issues with Wayland/Xwayland (more on that later), but the final verdict is very positive : Pi is nice and useful as a desktop environment, the hardware looks stable and the software dependable (but it is too early to say), and at 3W power consumption at idle (and 12W under full load), it will make the university happier. Actually it is so nice and the desktop experience so smooth and responsive, that I am seriously thinking about moving it to my desk at the office.
Hardware things
Not much to say here. The things I should mention are : (a) The Pi’s micro HDMI port is a bit difficult to work with, and you have to think whether you want to buy an adaptor or a fixed cable. (b) The official Pi.com case is not very good at cooling the Pi, maybe the official ‘active cooler’ is better and cheaper. There are also these ‘cases’ with double heat sinks and double fans for the fanatics. (c) It would be nice if your monitor comes with speakers, otherwise you will need either a USB- or a bluetooth-based set of speakers/headset/earbuds. (d) The ‘disk’ is a microSD card. My understanding is that this is the weak link hardware-wise. MicroSDs are not made for 24/7, you probably need a high-end model and properly scheduled backups to be on the safe side.
Software things
The Pi comes with Debian 12 (Bookworm), so everything works out of the box, except Wayland/Wayfire. I suffered considerably with the move away from X11, but in retrospect most of it was due to the complete absence of any familiarity from my side with the configuration of wayland and friends. Eventually we did reach an equilibrium and I am happy (for now ?), but note that some things like camera support and video-conferencing have not been tested. The Xwayland part (offering support for pure X11, non-wayland, apps) is not exactly a true substitute for a proper X11 server, and I even ended-up re-writing some code for two of my ancient programs which were using Ygl (an IrisGL-like X11-based substitute for GL) to make them run as fast as they do under X11. Having said all that, I do not regret staying with wayland : I learned things, and wayland appears to be faster and smoother and ‘fancier’ that X11. There are things that bug me, like the absence of a focus-follows-mouse setting (you need to find/build the extra plugins for wayfire to make this work), but all-in-all it looks good now. I suppose that someone with fewer preconceived ideas and expectations could have used the defaults happily and never experience any issues, but this was definitely not the case with me.
Tests in a ‘production’ environment
The Pi passed what I consider to be an acid test for a new desktop machine : log-in onto it, and try to work normally while using it for dealing with whatever happens to come along during a typical day at the office. I have been happily using it for several days now, and only occasionally did something needed to be installed/configured. The general feeling I got is that of using a normal fully-setup modern linux machine on a hardware that is up to the task.
 |
| A workspace with too many windows open. This is from an analysis performed remotely via ssh. |
Cost estimates
It looks like you are better off buying a kit and then add to it if needed, instead of buying the parts one-by-one. For example : buying separately the 8G model + the official Pi case + the official power supply costs 123.90 euros (incl. VAT). Buying this kit costs 124.90 euros, and you get (for the same price) the 8G model + the official active cooler + the official power supply + a HDMI ⇒ MicroHDMI cable + the (cheap-ish) ABS case + a Sandisk Ultra 32GB MicroSD (with pre-installed the OS). If 32G is not enough, you only have to buy a larger/better MicroSD card, attach a keyboard and a mouse, and you are good to go.
So, even if I add 30 euros for an extra MicroSD card + a set of wireless keyboard/mouse, we reach a total of ~155 euros for a complete ready-to-go Pi box. Add ~90 euros for a cheap 23" monitor with speakers, and you end-up with a grant total of ~250 euros for a complete Pi-based student terminal. Now : For 190 euros, you can buy a cheap chromebook with 11" display, two cores, and a 32G disk. But I can hardly imagine running e.g. pymol on a 11" display. I am writing this on a chromebook in vim and it is fine for that usage. But the chromebook is not a suitable device for molecular graphics, or having 5 windows open from an analysis of a molecular dynamics trajectory. To conclude this section : if you already have the monitor plus keyboard/mouse —and the 32G MicroSD is acceptable— you can buy a proper desktop machine with 125 euros, which is probably unbeatable. Even if you have to go for the full set, I doubt that you could find an as smooth and responsive machine with 250 euros.
Conclusions
The Raspberry Pi 5 is not a toy-like hardware suitable only for kids learning computing. It has sufficient resources to make a useful everyday terminal for college students as well. The Pi is ideal for writing code, testing it, and then deploying it to the servers for the production run. With the 8G model, you can easily have 4 workspaces loaded with a browser, a local tmux session, a remote session with X11 apps running an analysis, a local diary/journal for taking notes, etc., and with all those apps running, the responsiveness of the Raspberry remains essentially unaffected. You will, of course, not do your heavy computing on the Pi, but this is also true for all terminals we currently use. And with a price tag of 97€ for the 8G model, replacing a broken terminal comes as close to a walk in the park as you’ll ever get. Add the 3W power consumption at idle, and there you have it : a functional solution for light academic computing that is also light on the budget and also light with its environmental footprint. The way I see it, the only thing remaining to be seen concerns the robustness of the Pi : our classical PC-based student terminals can easily pass the 5 year mark, and we have several that have passed the 10 year mark. For the Pi to be useful, it should at least approach the 5 year mark. Replacing MicroSD cards every couple of months would definitely take the fun out of it. Time will show …
Update [April 2025] : Not only the Pi is still my main desktop machine (and I'm happy with it), but we have actually moved ahead and installed three devices for the students' office. The photo says it all :
Friday, September 29, 2023
A fully useable academic laptop for 189 € ? My take on a dirt-cheap chromebook.
I decided to write a review about this device¹ mainly because it pleasantly surprised me. My 5-year-old linux laptop took a hard fall, the casing broke, and I felt that to be on the safe side, I needed a ready-to-go backup device for this semester's teaching classes. I basically needed a device with a useable unix shell, an editor, gcc, a pdf viewer, and to be able to connect to the departmental projectors without issues. I stumbled across this chromebook by chance (actually, by sorting on the laptops' prices), and it caught my eye. I was always curious about what google is doing with chromeos and chromebooks, but my expectations from a 189€ chromebook with a weak CPU, 4G of physical memory, and only 32G disk space, were low. Extremely low. And I was wrong. This is a fully useable laptop for normal everyday academic applications. Having 4 workspaces ("desks") with dedicated email, unix shell, browser, a video call, and with all apps running simultaneously and being responsive and useable is the norm and not the exception. Add the ~10 hours of battery life on a single charge, the immediate availability of a proper linux (debian) distribution, the appealingly minimal, functional and stable chromeos environment, and it doesn't take long before you start thinking about taking it with you every time you leave home. And with a weight of 1.1 kgr, you might as well take it.
Don't get me wrong : you won't load this device with your brand new 10 Gbytes data set, and you will certainly not analyze the data with its AMD A6 processor. And you almost certainly won't do any short of serious photo/video editing or gaming on this chromebook either. But you can easily do all teaching presentations, all emailing, all everyday administrative tasks, right from this device.
However, do bear in mind that I'm an old guy. So old, that the following caveats apply :
Thursday, June 22, 2023
A new fat node has been added to the cluster. This is based on an Intel Core i7-13700KF (1700/3.4 GHz/30 MB), an NVIDIA MSI RTX 4070 Ti VENTUS 3X 12Gb OC 3, 8GB DDR4@3200 MHz, and two 2TB disks in a RAID1 array.
Initial tests confirmed that this is our new fastest machine, delivering with gromacs and HMR ~3930 ns/day on the FipWW 8,000 atom/5fs system. As with the other machines, with smaller systems the performance flattens at approximately ~4.1 μs/day (again with gromacs & HMR).
Saturday, May 13, 2023
The joy of WSL2
And just like that, there is no reason to fight with building native windows executables any longer. Install WSL2 on win11, and be done with it : all Linux graphical apps work out of the box, and at near native speeds. The screenshot below is from a molecular dynamics simulation analysis with carma / grcarma / stride / rasmol / evince / ..., all visibly working.
Monday, April 5, 2021
Martini & Gromacs & VMD & Bendix (and ROP)
Have been playing with the Martini force field for some time now. Still digging. Here is a short movie of a ROP mutant using Martini & Gromacs, and visualized with VMD & Bendix.
Friday, December 4, 2020
Just came out ...
«A molecular dynamics simulation study on the propensity of Asn-Gly-containing heptapeptides towards β-turn structures: Comparison with ab initio quantum mechanical calculations.»
Friday, February 14, 2020
New node added to the cluster
Initial tests confirmed that this is our new fastest machine, delivering ~575 ns/day on the standard 10,000 atom/4fs benchmark. As with the other i9, with smaller systems the performance flattens at approximately ~700ns/day with a 2.5fs step.
Monday, December 30, 2019
Illustrating transitions between conformers using circos plots
A new tutorial is available for preparing a circos plot illustrating the transitions between stable conformers identified from e.g. a folding molecular dynamics simulation.
Friday, July 26, 2019
Switching to a tiling window manager (i3)
Switching over to i3. First impressions are good : much less mouse herding, easier to focus, much lighter on resources, no bloating. Few issues with programs whose windows must remain floating (like the ccp4i stuff), but they are fixable through some elementary configuration. All-in-all this is very promising.
Even with tiled windows, ccp4i is still functional (and definitely more tidy) :
Tuesday, June 18, 2019
Fish & FZF
The combination of fish shell with fzf is outstanding. For example, you mistype a filename, hit <CTRL-T>, fzf finds it, and then with <CTRL-S> you can also examine the contents (with syntax highlighting with bat). Add z, fd, bd and rg to the soup and you have a whole new shell.
Friday, June 14, 2019
Short‐range periodicities paper.
Just came out : "On the presence of short‐range periodicities in protein structures that are not related to established secondary structure elements", https://doi.org/10.1002/prot.25758
Saturday, March 23, 2019
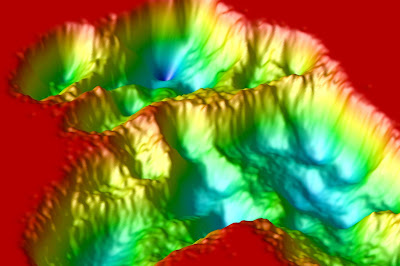
OpenDX rendering of folding landscapes
Folding landscape (from dPCA) of the M2TM peptide in TFE, rendered with OpenDX. Too much red probably.
Saturday, February 16, 2019
New fat box arrived
A new fat node has been added to the cluster. This is based on an Intel i9-9900K, a GTX-1080, 8GB DDR4@3000 MHz, and a two-disk RAID 0 @ 4TB.
Initial tests confirmed that this is indeed our new fastest machine, delivering ~560 ns/day on the standard 10,000 atom benchmark (second best is this one with 460 ns/day). With smaller systems, however, the performance flattens (giving with the CLN025 system 570 ns/day@2fs, ~730 ns/day@2.5fs).
Friday, February 8, 2019
Molecular simulation of peptides review
Just came out : "Molecular simulation of peptides coming of age: Accurate prediction of folding, dynamics and structures"